Import From Anywhere
Your files, NCBI, AddGene, SnapGene, iGEM…
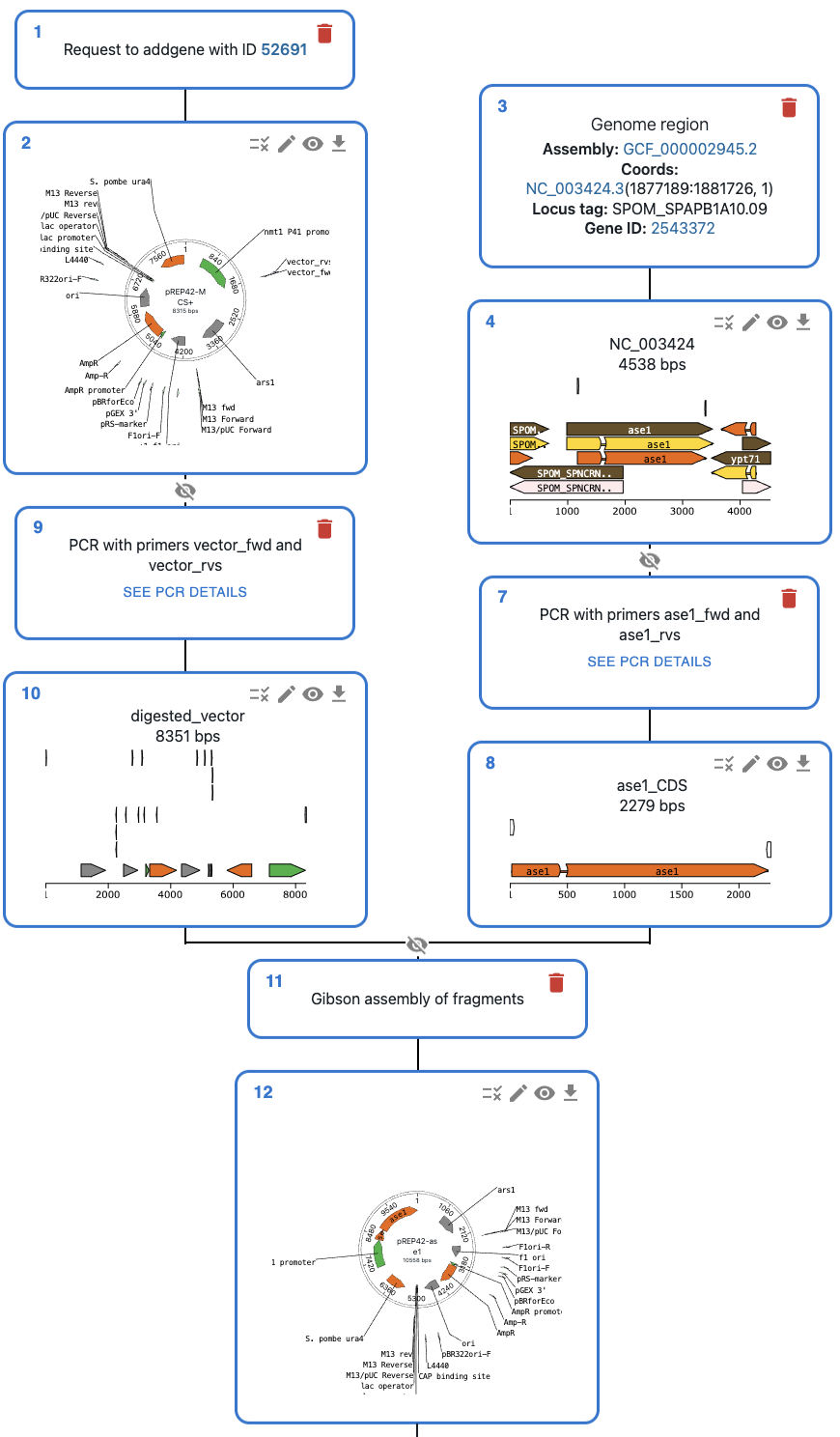
Engineer Anything
Gibson, Golden Gate, Gateway, In vivo assembly…
Automate Everything
Create scripts, notebooks, templates and forms
Share With Anyone
Export cloning workflows in FAIR Open File Format.
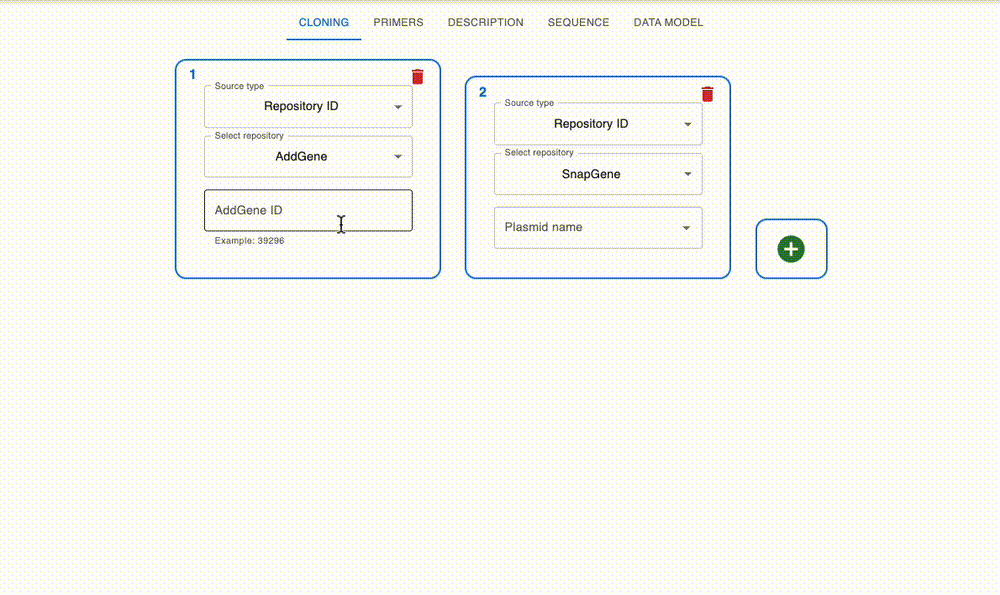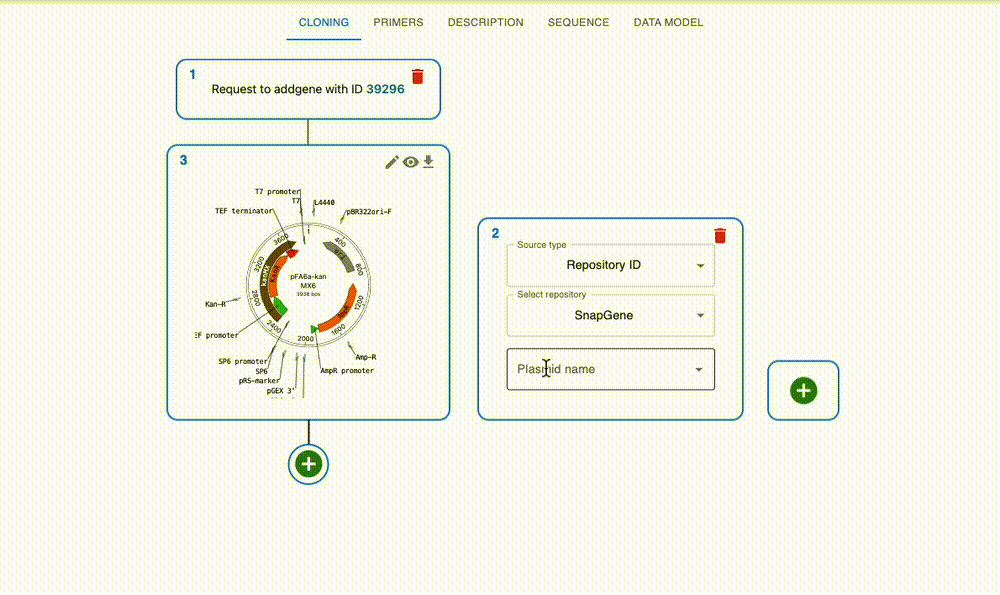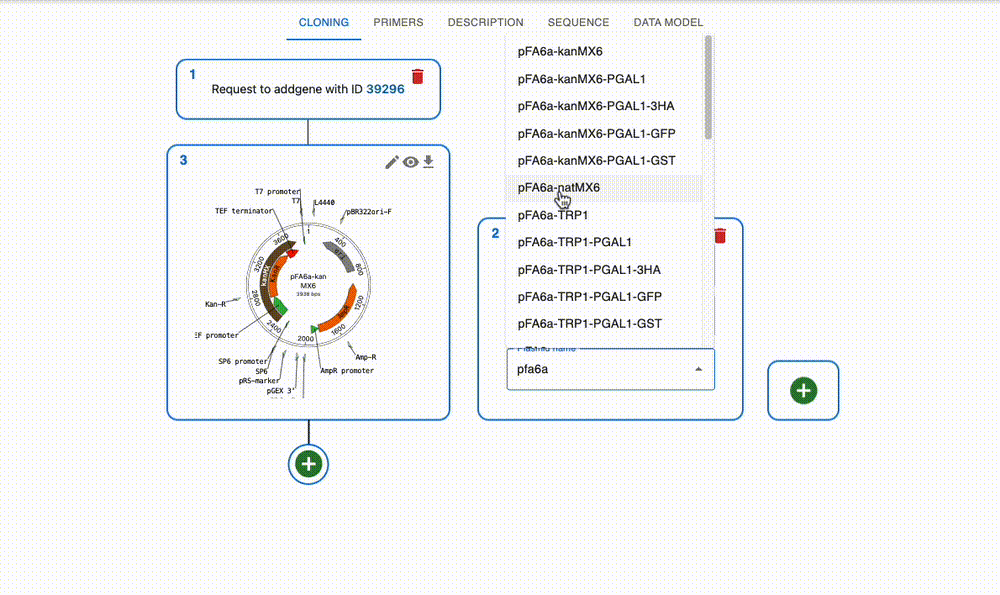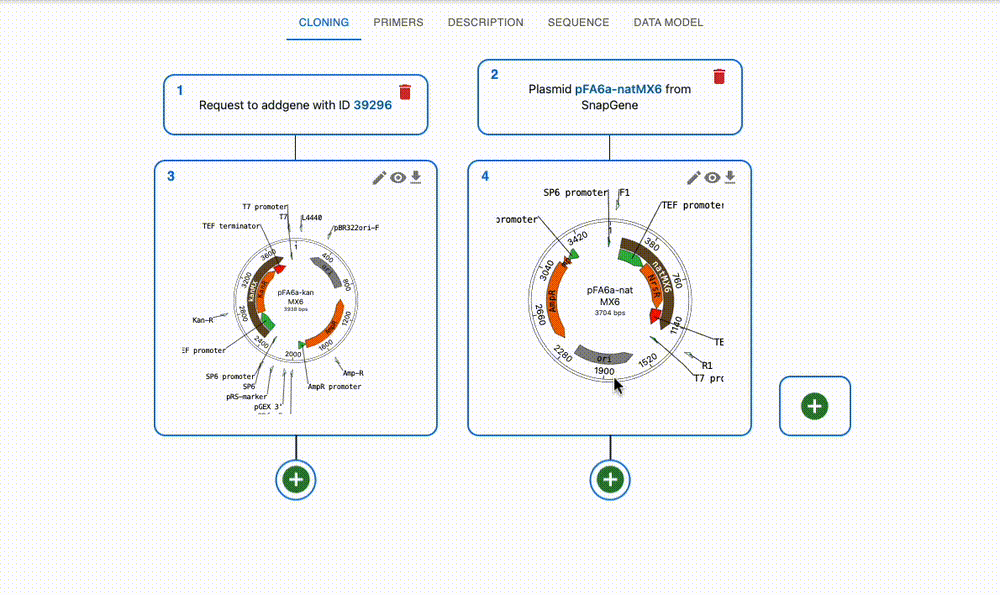
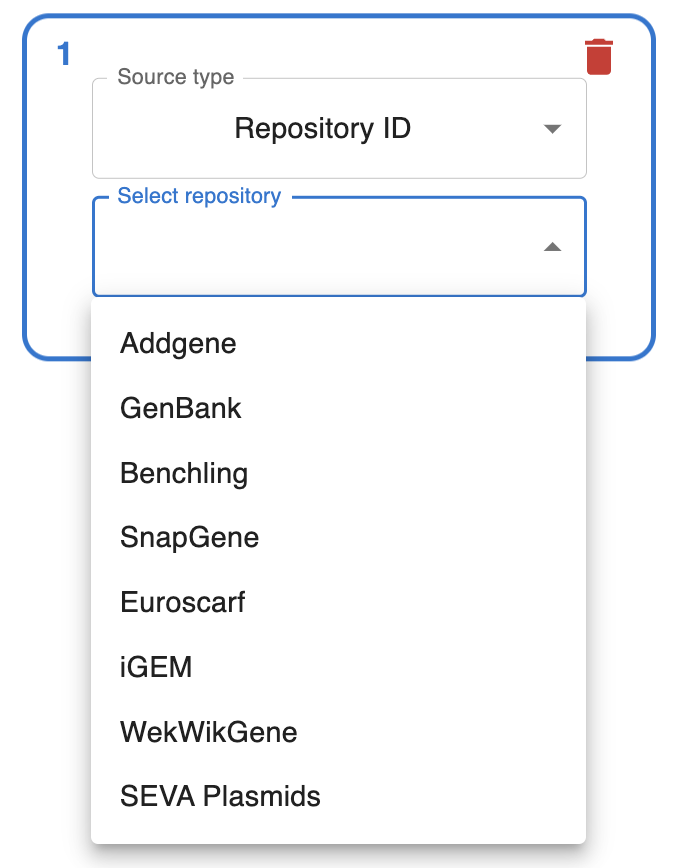
Import from anywhere
From many online repositories and local files.
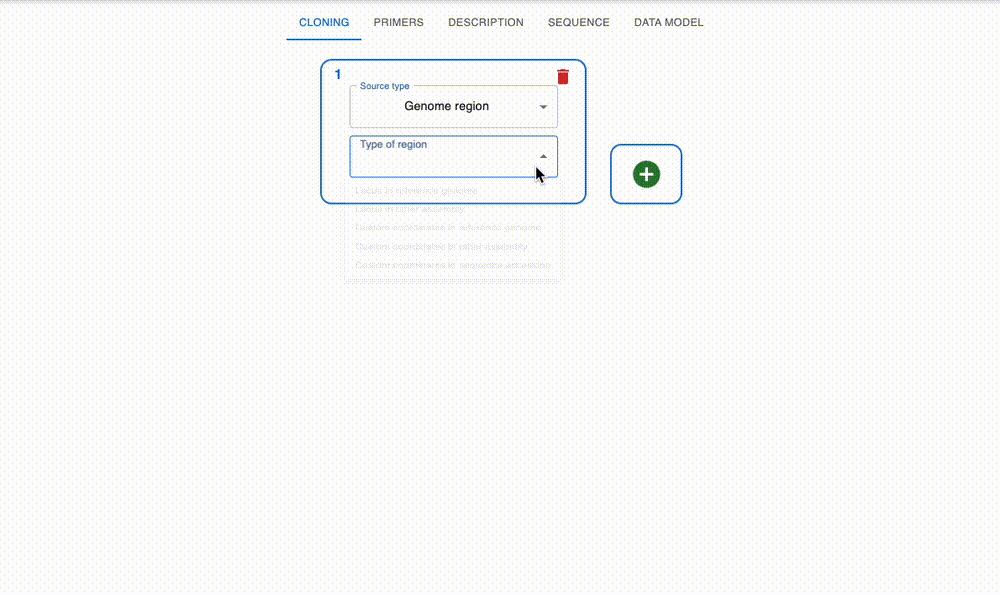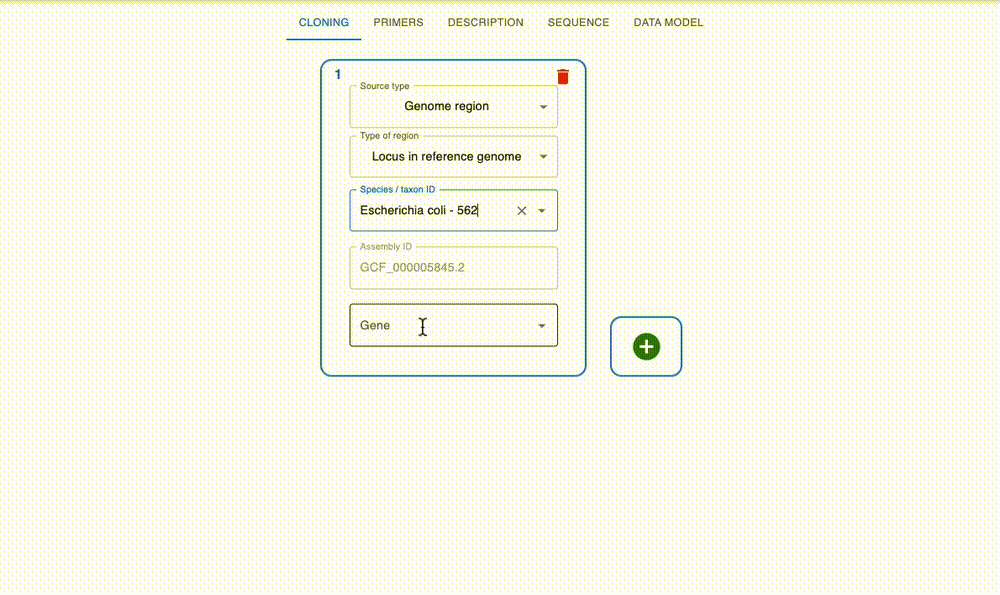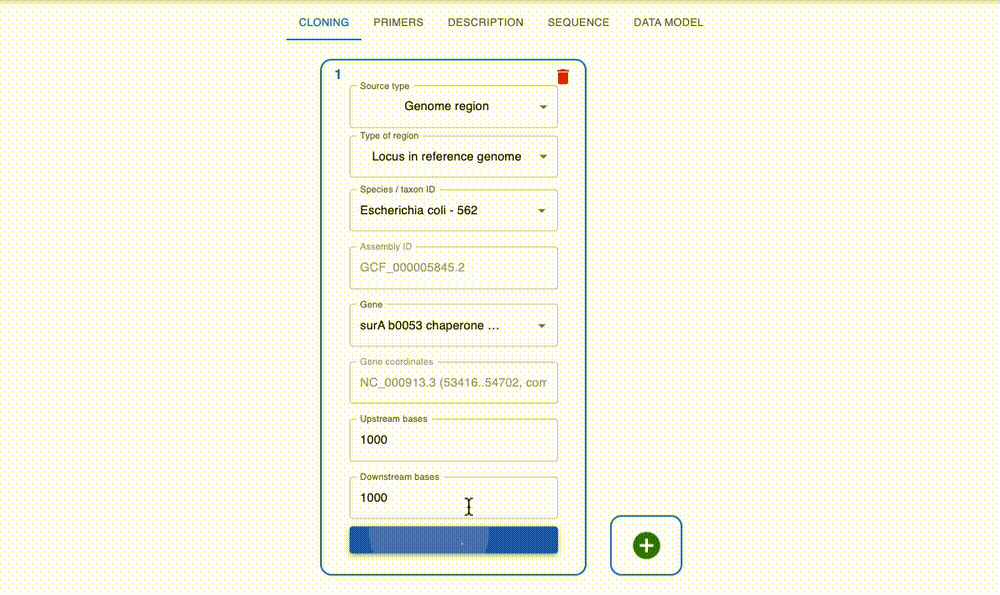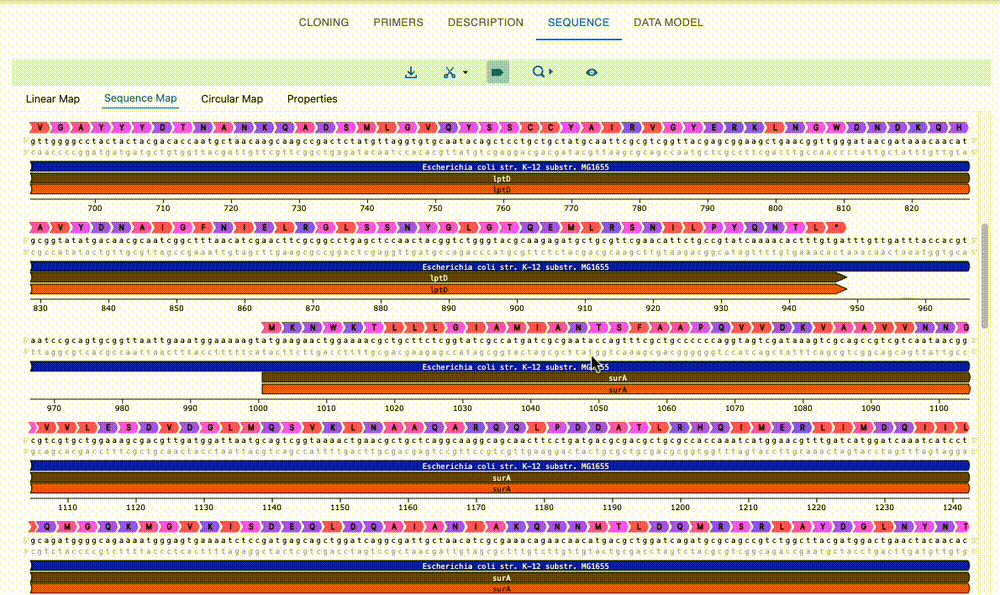
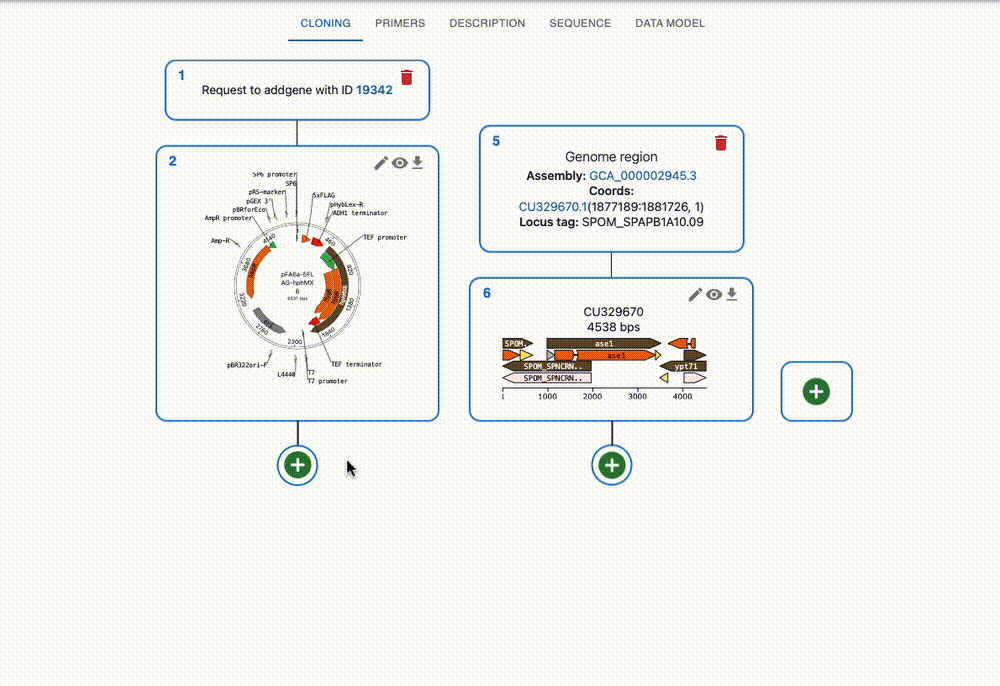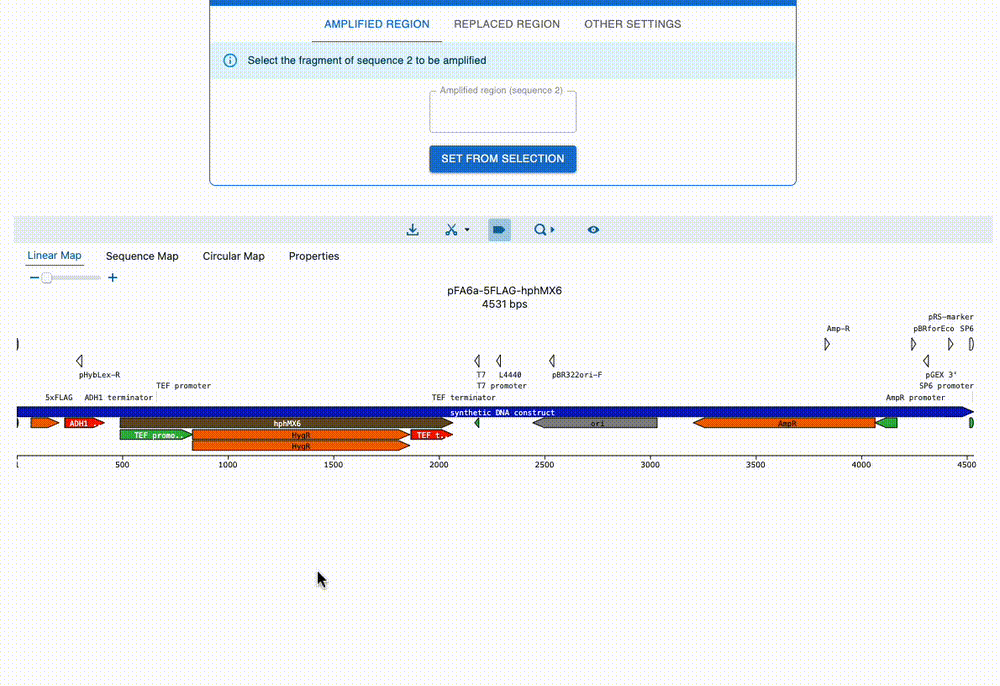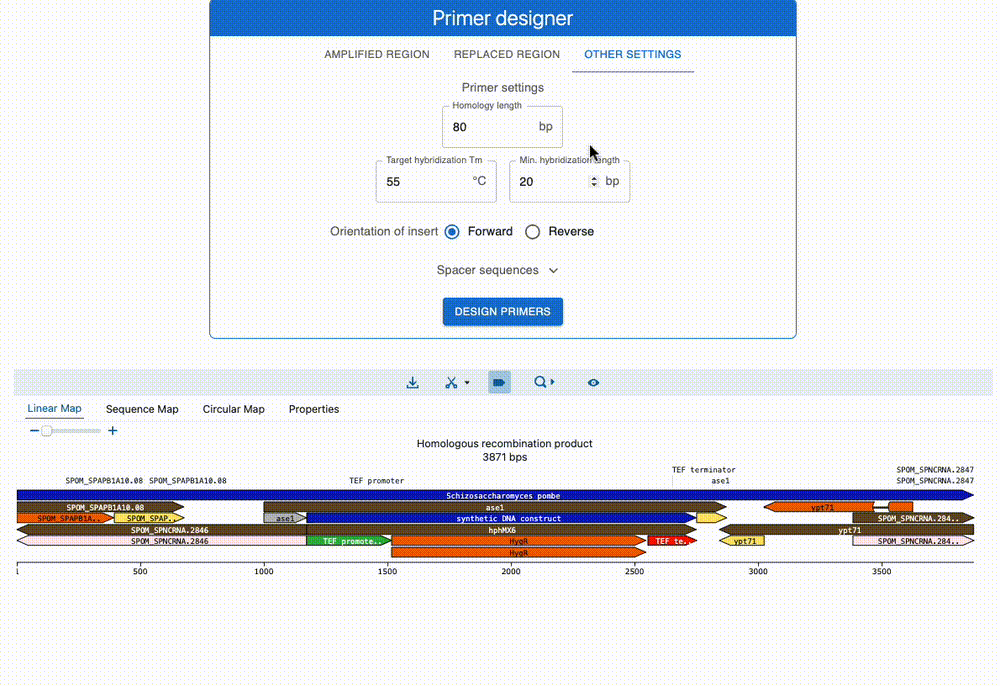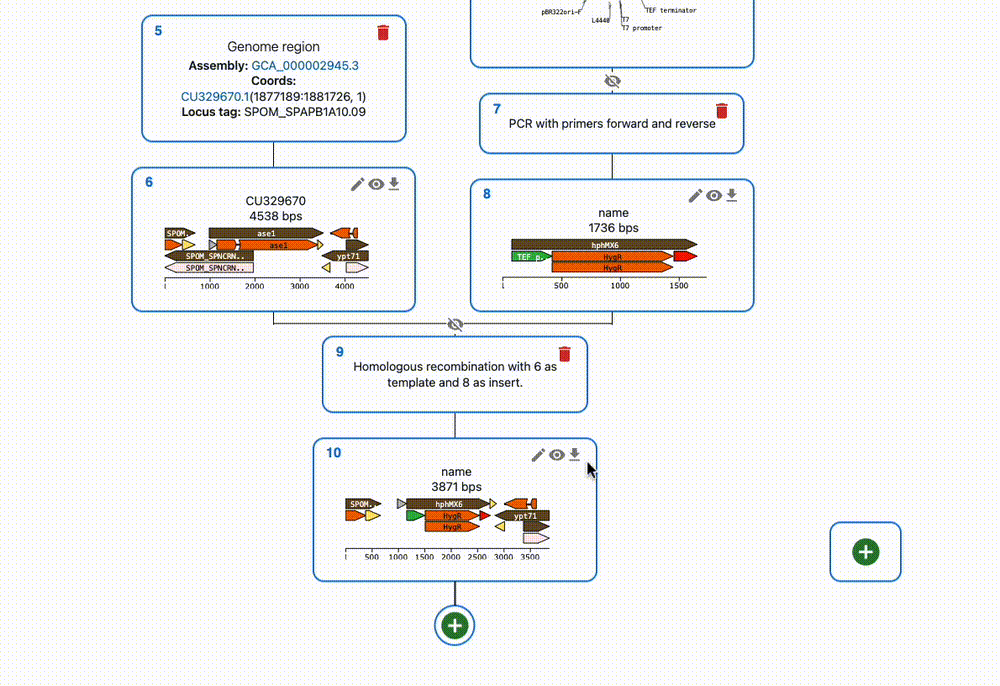
Engineer anything
Supports most cloning methods and has powerful primer design capabilities.
Automate everything
Use scripts, notebook, forms and templates to speed up design
Get started with scriptingStart using OpenCloning
Whether you work as a researcher, platform scientist, or in industry, OpenCloning can help you with DNA engineering.